iPRG Study Results
This year’s ABRF iPRG study “focuses on the evaluation of proteomics laboratories in identifying modified peptides present in a complex mixture.” Researchers were invited to analyze a common LC-MS/MS dataset, and report both the unmodified and modified peptides found in the data. PEAKS achieved outstanding performance in terms of both the total number of correctly identified peptides, and the number of correctly identified modified peptides that were spiked-in the sample.
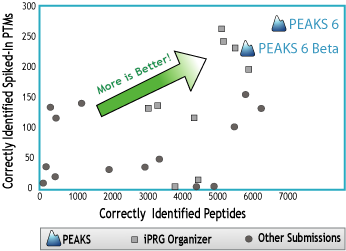
Figure 1: Scatter Plot depicting Peptides Correctly Identified vs Spiked-In PTMs Correctly Identified
This is another public study demonstrating that PEAKS is an excellent tool for large-scale in-depth proteomics analysis. PEAKS integrates four different algorithms for identifying database peptides, PTMs, mutations, and novel peptides, respectively. This in-depth analysis of your LC-MS/MS data ensures that no peptide is left unidentified.

