
SILAC Quantification
Overview
Stable isotope labeling by amino acids in cell culture (SILAC) is a powerful and popular approach for mass spectrometry (MS)-based quantitative proteomics
Highlights
- Accurate and sensitive detection and association of SILAC feature pairs
- Transfer IDs between associated SILAC pairs and different MS runs
- Flexible experimental design and statistical tools to facilitate data analysis
Introduction of SILAC and Super-SILAC
SILAC has become one of the most popular labeling techniques for MS-based quantitative proteomics [1]. In this metabolic labeling strategy, differential isotope labeled samples (proteins/peptides) are combined early in the experimental procedure and analyzed together by LC-MS/MS. Therefore, the variation introduced from sample processing is minimized. Since stable-isotope labeled peptides have almost the same physicochemical properties as their natural counterparts, the same peptides with differential labeling co-elute from the liquid chromatography (LC) column and their amounts can be accurately quantified relative to each other.
An extension of this labeling strategy is named super-SILAC [2, 3], where labeled samples can be produced separately and spiked into each of the experimental samples that are not amenable to metabolic labeling, e.g. human tissues. This pool of spiked-in heavy proteins is used as an internal standard for quantification.
PEAKS Q supports analyses of SILAC and super-SILAC types of data.
SILAC Quantification Algorithms in PEAKS Q
- ID transfer between associated SILAC pairs
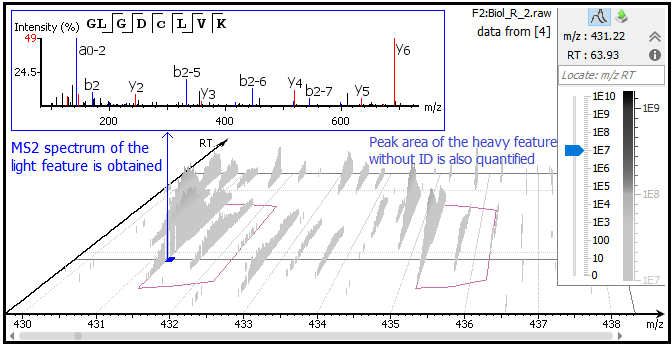
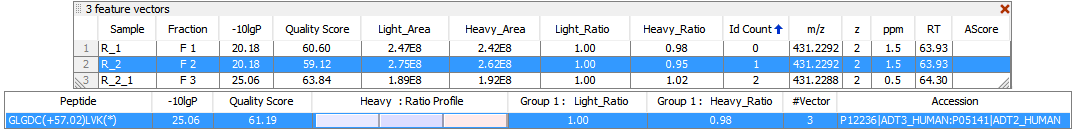
PEAKS Q detects and associates 2- or 3-plex SILAC feature pairs that have the same charge, similar MS1 peak area correlation over retention time, expected mass shifts caused by labeling and fall within certain mass errors. If an identification is obtained from one of the labeled states, the whole SILAC pair feature can be quantified and used for peptide and protein ratio calculations. For example, the light form of peptide GLGDCLVK was fragmented and identified in sample R_2 [4]. Although no MS2 spectrum was obtained from the K8 labeled heavy counterpart, the SILAC pair could still be quantified in PEAKS Q as highlighted in the feature vectors table. “Id Count” indicates the number of MS2 identified for each SILAC pair.
- ID transfer between different MS runs after alignment
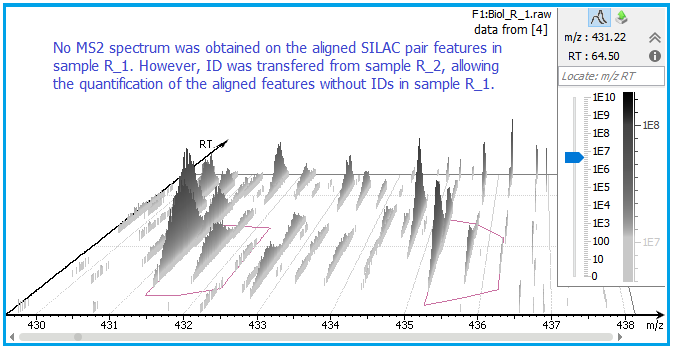
Retention times of different LC-MS runs are first aligned, then the MS/MS and ID can be matched from another run by aligning features within tight mass ranges and retention times, allowing quantification of SILAC pairs without any ID.

References
- Ong, S. E., Blagoev, B., Kratchmarova, I., Kristensen, D. B., Steen H., Pandey A., Mann M., Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics, 1, 376-386 (2002). https://doi.org/10.1074/mcp.M200025-MCP200
- Neubert, T. A., Tempst, P., Super-SILAC for tumors and tissues. Nat. Methods, 7, 361-362 (2010). https://doi.org/10.1038/nmeth0510-361
- Geiger, T., Cox, J., Ostasiewicz, P., Wisniewski, J. R., Mann, M., Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods, 7, 383-385 (2010). https://doi.org/10.1038/nmeth.1446
- Liberski, A. R., Al-Noubi, M. N., Rahman, Z. H., Halabi, N. M., et al., Adaptation of a commonly used, chemically defined medium for human embryonic stem cells to stable isotope labeling with amino acids in cell culture. J. Proteome Res., 12, 3233-3245 (2013). https://doi.org/10.1021/pr400099j
Resources
- Application Note 1: Single-group SILAC-based data analysis in PEAKS Studio. [Download PDF]
- Application Note 2: Multiple-group SILAC-based data analysis in PEAKS Studio. [Download PDF]
- Application Note 3: SILAC-based PTM profiling analysis in PEAKS Studio. [Download PDF]

