
 Ile/Leu Differentiation
Ile/Leu Differentiation
Highlights
- Design and perform LC-MS/MS analysis (≥50μg of a purified protein)
- Guarantee accuracy by multiple proofs
- Guarantee two-week turn-around and service quality
Why is Ile/Leu differentiation needed?
Although isoleucine (Ile) and leucine (Leu) share exactly the same molecular weight, their different structures may dramatically influence protein functions. To thoroughly characterize a protein of interest, especially a therapeutic protein, researchers have to cautiously examine each Ile/Leu position in the protein sequence. Particularly, for Ile/Leu residues in and near CDRs of an antibody, flipping between Ile/Leu may result in significant differences in its biochemical properties.
Our Ile/Leu differentiation service provides a precise identification of Ile and Leu residues during protein sequencing and to help dramatically reduce the cost in the subsequent expression stage. For instance, a target protein which contains 4 undetermined Ile/Leu residues would need up to 16 constructs expressed to fully test which residue is correct. This does not even take codon combinations into account! With our Ile/Leu differentiation service, you no longer need to guess and repeatedly test, but be 100% certain from the start.
For the purpose of protein sequence validation, our Ile/Leu differentiation service can help you confirm the expression of Ile/Leu in your protein of interest and discover potential sequence variants.
How to Differentiate Ile/Leu?
- Ile and Leu residues are generally considered to be indistinguishable by traditional MS. However, w-ions of isoleucine and leucine generated by certain MS methods can distinguish them 1,2,3;
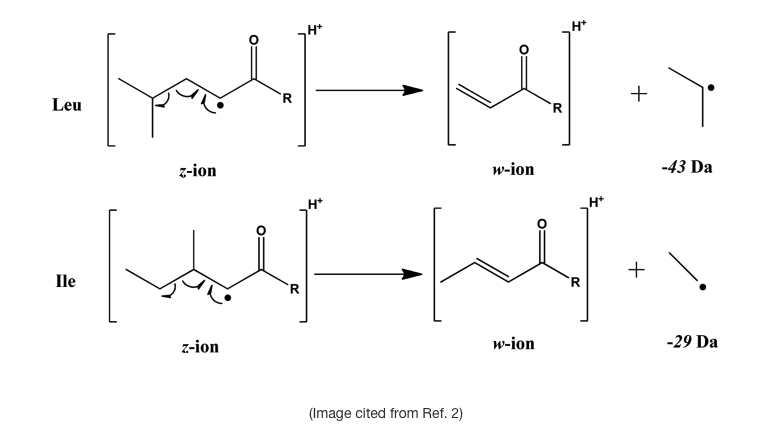
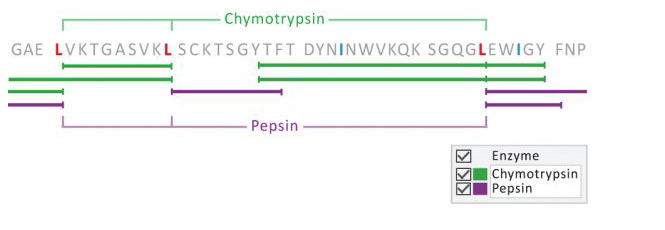
- (Optional) The use of enzymes is important as a diagnostic and research tool because of the specificity they exhibit. By exploiting enzymatic cleavage rules, we can use enzymes such as chymotrypsin or pepsin, for example, to differentiate between Ile/Leu residues;
- (Optional) Statistical comparison of homologous databases for conservative sites in the mAb constant regions and framework regions.
- Johnson, R. S., Martin, S., Biemann, K., Stults, J. T., & Watson, J. T. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal Chem. 1987 Nov 1;59(21):2621-5.
- Xiao, Y., Vecchi, M. M., & Wen, D. Distinguishing between Leucine and Isoleucine by Integrated LC-MS Analysis using an Orbitrap Fusion Mass Spectrometer. Anal Chem. 2016 Nov 1;88(21):10757-10766.
- Zhokhov, S. S., Kovalyov, S. V., Samgina, T. Y., & Lebedev, A. T. An EThcD-Based Method for Discrimination of Leucine and Isoleucine Residues in Tryptic Peptides. J Am Soc Mass Spectrom. 2017 Aug;28(8):1600-1611.

