Article: Green Daniel R., et al, Mapping the Tooth Enamel Proteome and Amelogenin Phosphorylation Onto Mineralizing Porcine Tooth Crowns, Frontiers in Physiology. V. 10, 2019, 295. DOI: 10.3389/fphys.2019.00925
Abstract
Tooth enamel formation is important for strong, healthy teeth and for the prevention of dental diseases. So, understanding the regulatory mechanisms involved in enamel formation is important. Daniel Green and his coauthors recently published, “Mapping the Tooth Enamel Proteome and Amelogenin Phosphorylation Onto Mineralizing Porcine Tooth Crowns” in Frontiers in Physiology. Through knowledgeable use of PEAKS Studio PTM Profiling capabilities, they were able to characterize the involvement of protein phosphorylation of enamel matrix proteins at different stages of development. Their label-free quantification analysis through PEAKS Studio also showed greater protein diversity than was previously reported. This protein characterization approach can be applied to any research where any understanding of different amounts of proteins and protein modifications is needed.
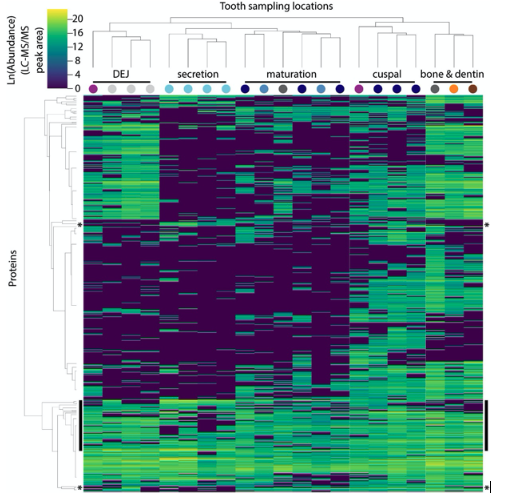
Hierarchical clustering of peptides and sample locations based upon abundance patterns. Overview of all 551 peptides and 21 sample locations clustered by Euclidian distance, showing that secretory proteins are tightly clustered compared to other sampled locations, a result consistent with our PCA (Figure 1B). Sample location clustering compared to a priori anatomical assignment indicated by colored circles (see Figure 1). Most known amelogenesis proteins including P173+LRAP products, AMBN, ENAM and MMP20 cluster tightly together (vertical black bars); stars indicate the location of the P190 AMELX isoform (above) and KLK4 (below).

