Featured Article:
Abstract
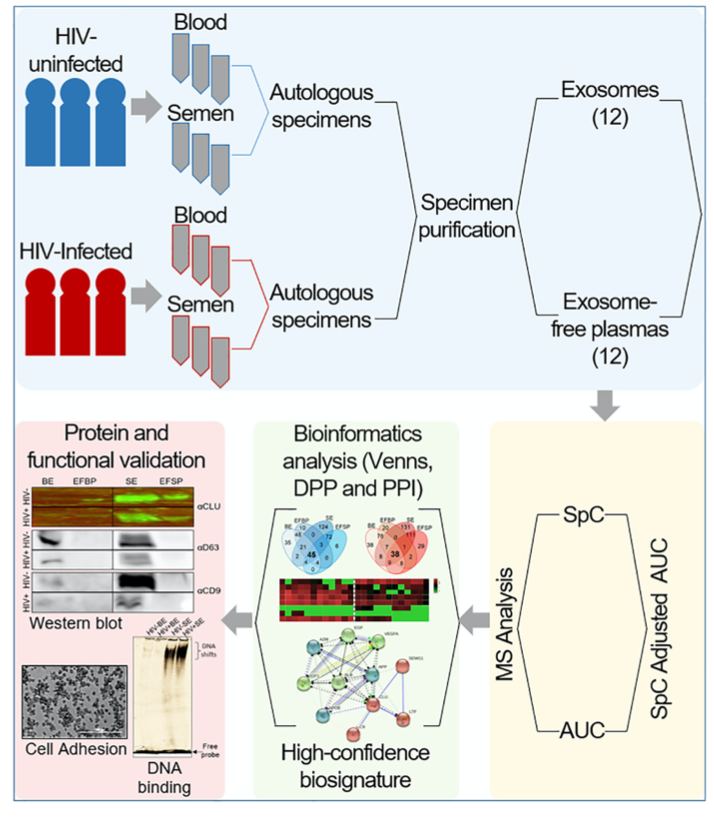
In Kaddour et. al’s recent publication in the American Society for Biochemistry and Molecular Biology, PEAKS Studio was used to identify and quantify proteins found in HIV- and HIV+ participants’ blood and semen. A workflow was designed for this comparative study called the Systematic Multifactorial Proteomic Profiling (SMPP) to specifically control for errors and noise. Samples are autologous to eliminate confounding factors from different donors. For the MS analysis, a combination of spectral counting (SpC) and area under curve (AUC) were used as an initial step to obtain high-confidence biosignatures.
A total of 587 proteins were found to be overlapping in the two protein lists at 1% FDR, -10lgP ≥ 50 and at least 2 unique peptides for the SpC method, and 105 a. u. cut-off for the AUC method. The label-free method was used to quantify the proteins, which visualized the difference in abundance between the blood and semen proteomes, as well as HIV- versus HIV+ samples. With the proteins identified and quantified using PEAKS, the authors hope to have their findings considered as biomarkers applicable for medicine.